FastSurfer - a fast and accurate deep-learning based neuroimaging pipeline
FastSurfer is a fast and extensively validated deep-learning pipeline for the fully automated processing of structural human brain MRIs. As such, it provides FreeSurfer conform outputs, enables scalable big-data analysis and time-critical clinical applications such as structure localization during image acquisition or extraction of quantitative measures.
FastSurfer consists of two main parts building upon each other:
-
FastSurferCNN - an advanced deep learning architecture capable of whole brain segmentation into 95 classes in under 1 minute, mimicking FreeSurfer’s anatomical segmentation and cortical parcellation (DKTatlas)
-
recon-surf - full FreeSurfer alternative for cortical surface reconstruction, mapping of cortical labels and traditional point-wise and ROI thickness analysis in approximately 60 minutes (+ optionally 30 min for group registration).
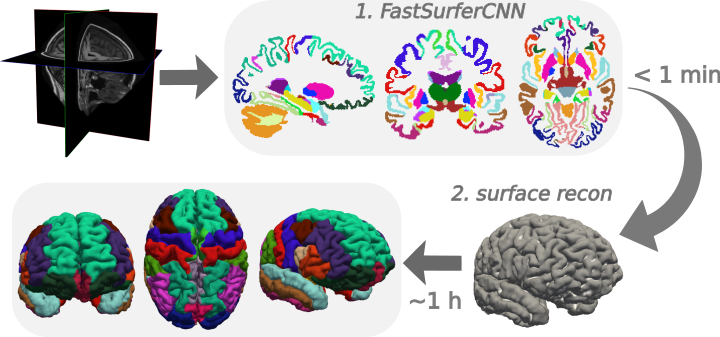
Fig 1. FastSurfer - fully automated neuroimaging pipeline combining an advanced neural network architecture (FastSurferCNN) with a surface pipeline (recon-surf).
Volumetric Segmentation - FastSurferCNN
FastSurferCNN is composed of three fully convolutional neural networks (F-CNNs) operating on coronal, axial, and sagittal 2D slice stacks and a final view aggregation combining the advantages of 3D patches (local neighbourhood) and 2D slices (global view). Within each F-CNN we incorporate local and global competition via competitive dense blocks and competitive skip pathways, as well as multi-slice information aggregation (7-channel input) that specifically tailor network performance towards accurate recognition of both cortical and sub-cortical structures.
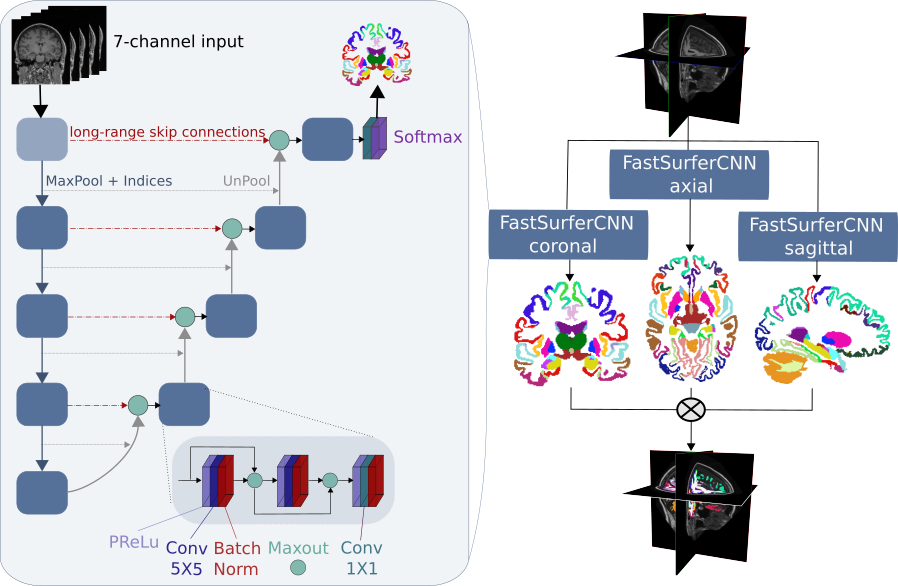
Fig 2. FastSurferCNN architecture and proposed view aggregation.
Surface reconstruction - recon-surf
With recon-surf we perform fast cortical surface reconstruction, mapping of cortical labels and traditional point-wise and ROI thickness analysis. Due to the presence of a high-quality brain segmentation (generated by FastSurferCNN) many steps from within the traditional FreeSurfer recon-all have become obsolete, such as skull stripping and non-linear atlas registration.
Further, we perform a novel spectral spherical embedding and directly map the cortical labels from the image to the surface. Precisely, we use the eigenfunctions of the Laplace-Beltrami operator to parametrize the surface smoothly and quickly generate the final spherical map by scaling the 3D spectral embedding vector to unit length.
Overall, these improvements result in a significant speed-up compared to FreeSurfer.
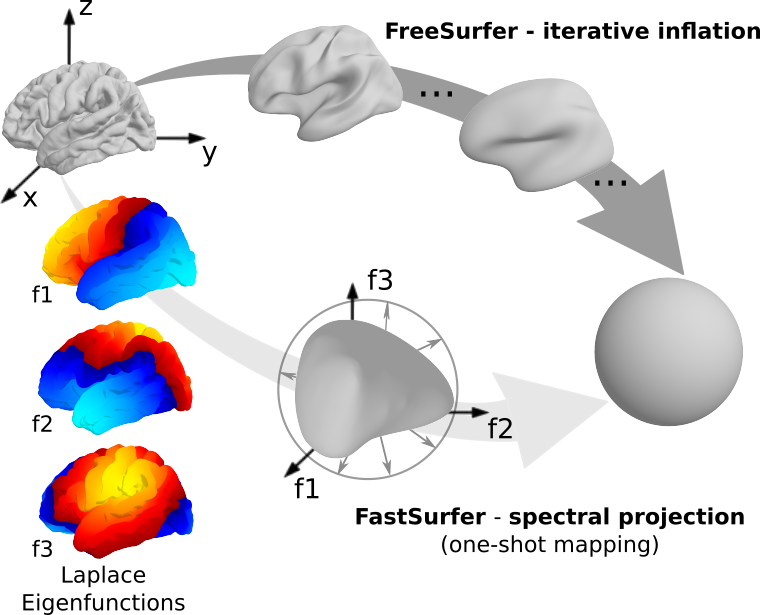
Fig 3. FreeSurfer´s iterative spherical inflation versus recon-surf´s one-shot spectral embedding using the first three non-constant eigenfunctions of the Laplace-Beltrami operator.
Proof of Concept
To ensure the usefulness of our entire pipeline, FastSurfer was extensively validated using a number of publicly available datasets.
1. Accuracy and Generalizability - How good is the whole brain segmentation generated by FastSurferCNN?
Answer: Very good! FastSurferCNN outperforms other deep-learning architectures by a significant margin both wrt to FreeSurfer and a manual standard (Mindboggle). It generalizes well across five different datasets including subjects with different disease states (e.g. cognitive normal, mild cognitive impaired or demented subjects in OASIS, ADNI, MIRIAD), different vendors (e.g. THP), age groups, downsampled and defaced images (e.g. HCP) and alternative T1-imaging protocols (e.g. MIRIAD)
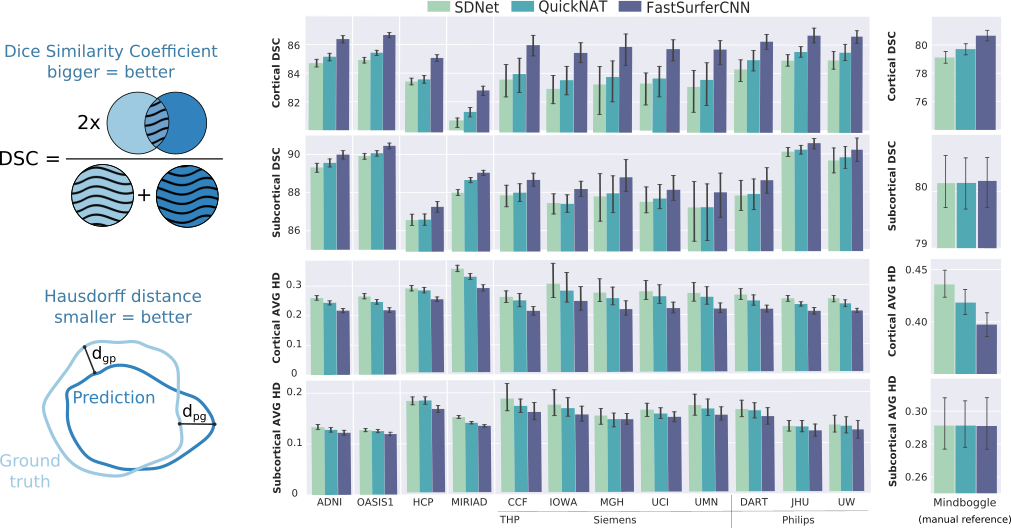
Fig 4. FastSurfer outperforms other state-of-the-art deep-learning architectures across a number of datasets both wrt to Dice Similarity Coefficient and Hausdorff distance.
2. Reliability - How well does FastSurfer reproduce results across scans with minimal anatomical variations (Test-Retest)?
Answer: FastSurfer is highly reliable as demonstrated by the close agreement between the thickness and volumetric measurements for 20 Test-Retest subjects from OASIS1.
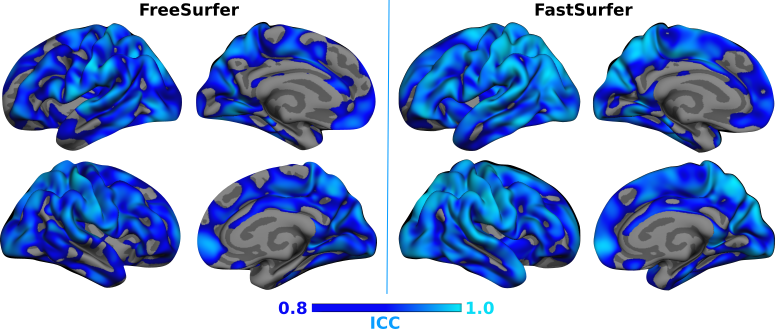
Fig 5. FreeSurfer exhibits high test-retest reliability and improves agreement between the cortical thickness measurments compared to FreeSurfer.
3. Sensitivity - How well does FastSurfer detect significant effects in the data?
Answer: FastSurfer is capable of accurately reproducing known disease effects in a control versus dementia cross-sectional group study. Reduced cortical thickness in regions associated with dementia (e.g. temporal lobes) as well as subcortical volume differences (e.g. enlarged ventricles, shrunken Hippocampus) are robustly detected with increased sensitivity relative to FreeSurfer.
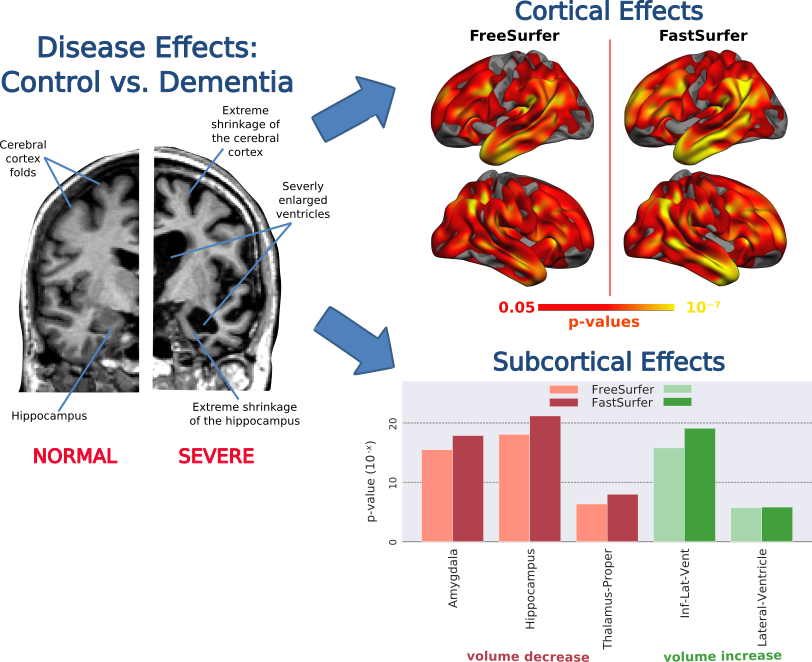
Fig 6. FastSurfer sensitively detects known differences in the cortical thickness and subcortical volumes between dementia and control groups.
Tool and Paper
-
FastSurfer is available as open source at github.
-
Documentation, usage and installation instructions are available here.
-
An accompanying set of quality control tools can be found at github.
-
In-depth information about FastSurfer can be found in our paper.
-
Or watch our small presentation (10min) on YouTube.
Tutorial
- A set of detailed tutorials for running Fastsurfer can be found within the FastSurfer github repository at github.
News
- FastSurfer was presented in the #OHBMx equinox (check out our twitter @deepmilab).